Quantitative Data¶
Feature¶
In OpenMS, information about quantitative data is stored in a so-called
Feature which we have previously discussed here. Each
Feature represents a region in RT and m/z space use for quantitative
analysis.
1 2 3 4 5 6 7 | from pyopenms import *
feature = Feature()
feature.setMZ( 500.9 )
feature.setCharge(2)
feature.setRT( 1500.1 )
feature.setIntensity( 30500 )
feature.setOverallQuality( 10 )
|
Usually, the quantitative features would be produced by a so-called
“FeatureFinder” algorithm, which we will discuss in the next chapter. The
features can be stored in a FeatureMap and written to disk.
1 2 3 4 5 6 7 8 | fm = FeatureMap()
fm.push_back(feature)
feature.setRT(1600.5 )
feature.setCharge(2)
feature.setMZ( 600.0 )
feature.setIntensity( 80500.0 )
fm.push_back(feature)
FeatureXMLFile().store("test.featureXML", fm)
|
Visualizing the resulting map in TOPPView allows detection of the two
features stored in the FeatureMap with the visualization indicating charge
state, m/z, RT and other properties:
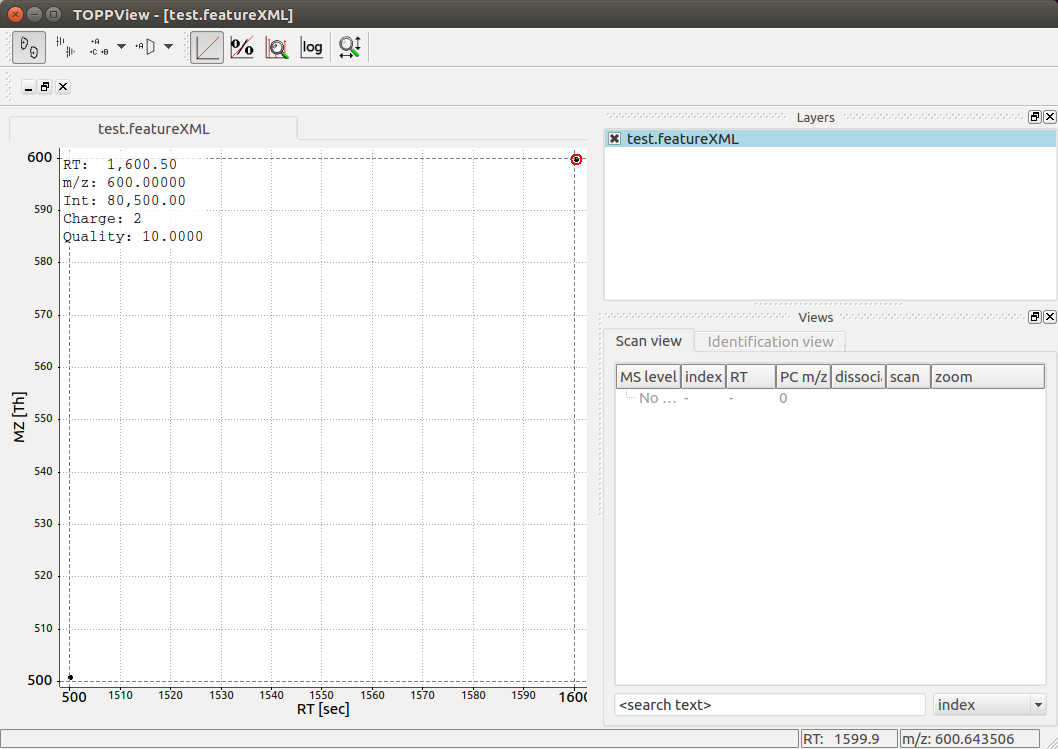
Note that in this case only 2 features are present, but in a typical LC-MS/MS experiments, thousands of features are present.
FeatureMap¶
The resulting FeatureMap can be used in various ways to extract
quantitative data directly and it supports direct iteration in Python:
1 2 3 4 5 | from pyopenms import *
fmap = FeatureMap()
FeatureXMLFile().load("test.featureXML", fmap)
for feature in fmap:
print("Feature: ", feature.getIntensity(), feature.getRT(), feature.getMZ())
|
ConsensusFeature¶
Often LC-MS/MS experiments are run to compare quantitative features across
experiments. In OpenMS, linked features from individual experiments are
represented by a ConsensusFeature
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 | from pyopenms import *
feature = ConsensusFeature()
feature.setMZ( 500.9 )
feature.setCharge(2)
feature.setRT( 1500.1 )
feature.setIntensity( 80500 )
# Generate ConsensusFeature and features from two maps (with id 1 and 2)
### Feature 1
f_m1 = ConsensusFeature()
f_m1.setRT(500)
f_m1.setMZ(300.01)
f_m1.setIntensity(200)
f_m1.ensureUniqueId()
### Feature 2
f_m2 = ConsensusFeature()
f_m2.setRT(505)
f_m2.setMZ(299.99)
f_m2.setIntensity(600)
f_m2.ensureUniqueId()
feature.insert(1, f_m1 )
feature.insert(2, f_m2 )
|
We have thus added two features from two individual maps (which have the unique
identifier 1 and 2) to the ConsensusFeature.
Next, we inspect the consensus feature, compute a “consensus” m/z across
the two maps and output the two linked features:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 | # The two features in map 1 and map 2 represent the same analyte at
# slightly different RT and m/z
for fh in feature.getFeatureList():
print(fh.getMapIndex(), fh.getIntensity(), fh.getRT())
print(feature.getMZ())
feature.computeMonoisotopicConsensus()
print(feature.getMZ())
# Generate ConsensusMap and add two maps (with id 1 and 2)
cmap = ConsensusMap()
fds = { 1 : ColumnHeader(), 2 : ColumnHeader() }
fds[1].filename = "file1"
fds[2].filename = "file2"
cmap.setColumnHeaders(fds)
feature.ensureUniqueId()
cmap.push_back(feature)
ConsensusXMLFile().store("test.consensusXML", cmap)
|
Inspection of the generated test.consensusXML reveals that it contains
references to two LC-MS/MS runs (file1 and file2) with their respective
unique identifier. Note how the two features we added before have matching
unique identifiers.
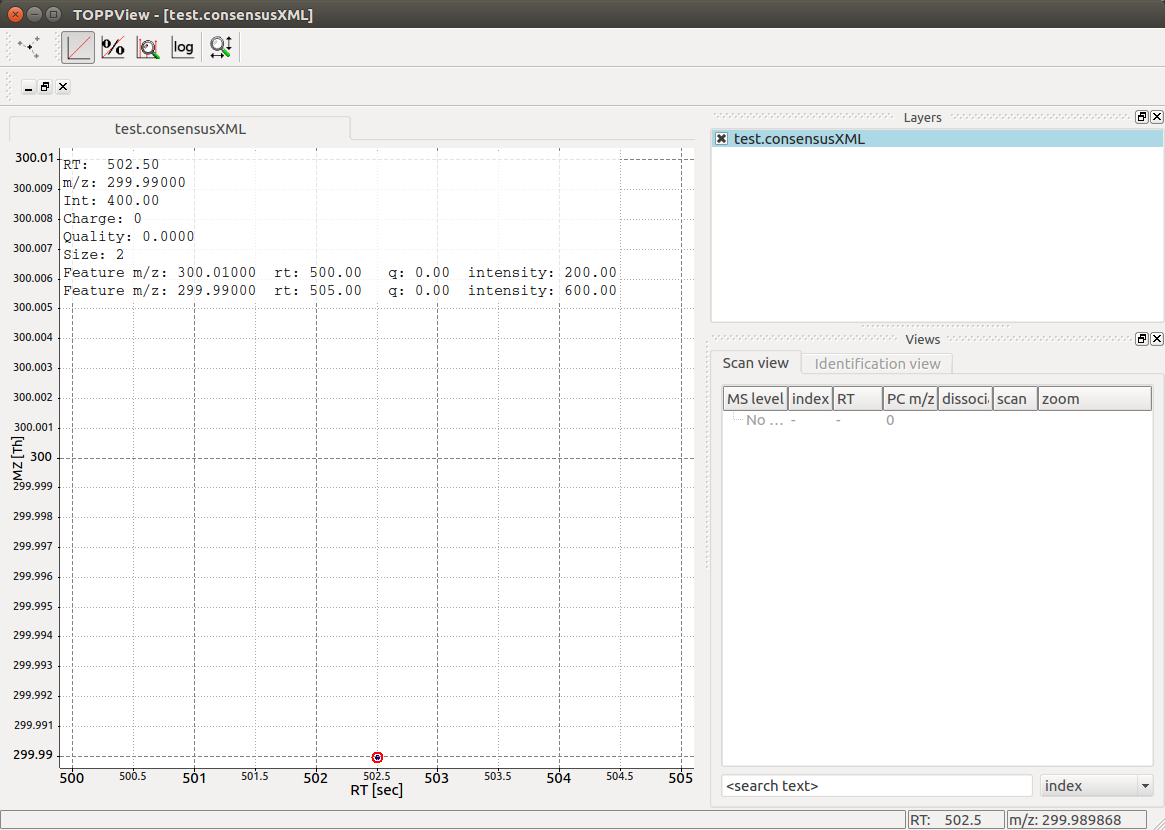
Visualization of the resulting output file reveals a single
ConsensusFeature of size 2 that links to the two individual features at
their respective positions in RT and m/z:
ConsensusMap¶
The resulting ConsensusMap can be used in various ways to extract
quantitative data directly and it supports direct iteration in Python:
1 2 3 4 5 6 7 8 9 10 | from pyopenms import *
cmap = ConsensusMap()
ConsensusXMLFile().load("test.consensusXML", cmap)
for cfeature in cmap:
cfeature.computeConsensus()
print("ConsensusFeature", cfeature.getIntensity(), cfeature.getRT(), cfeature.getMZ())
# The two features in map 1 and map 2 represent the same analyte at
# slightly different RT and m/z
for fh in cfeature.getFeatureList():
print(" -- Feature", fh.getMapIndex(), fh.getIntensity(), fh.getRT())
|