Interactive Plots#
With special plotting libraries like holoviews and datashader for big
data visualization as well as bokeh for interactiveness, we can use the
functionality of pyOpenMS to quickly create fully interactive views of
mass spectrometry data. Here we plot a full map of MS1 that can be
interactively zoomed-in if you execute the code in a notebook
(e.g. on Binder, see the button on top of the page).
1import pyopenms as oms
2import pandas as pd
3import numpy as np
4import datashader as ds
5import holoviews as hv
6import holoviews.operation.datashader as hd
7from holoviews.plotting.util import process_cmap
8from holoviews import opts, dim
9import sys
10
11hv.extension("bokeh")
12
13exp = oms.MSExperiment() # type: PeakMap
14loader = oms.MzMLFile()
15loadopts = loader.getOptions() # type: PeakFileOptions
16loadopts.setMSLevels([1])
17loadopts.setSkipXMLChecks(True)
18loadopts.setIntensity32Bit(True)
19loadopts.setIntensityRange(oms.DRange1(oms.DPosition1(5000), oms.DPosition1(sys.maxsize)))
20loader.setOptions(loadopts)
21loader.load("../../../src/data/BSA1.mzML", exp)
22exp.updateRanges()
23expandcols = ["RT", "mz", "inty"]
24spectraarrs2d = exp.get2DPeakDataLong(
25 exp.getMinRT(), exp.getMaxRT(), exp.getMinMZ(), exp.getMaxMZ()
26)
27spectradf = pd.DataFrame(dict(zip(expandcols, spectraarrs2d)))
28spectradf = spectradf.set_index(["RT", "mz"])
29
30maxrt = spectradf.index.get_level_values(0).max()
31minrt = spectradf.index.get_level_values(0).min()
32maxmz = spectradf.index.get_level_values(1).max()
33minmz = spectradf.index.get_level_values(1).min()
34
35
36def new_bounds_hook(plot, elem):
37 x_range = plot.state.x_range
38 y_range = plot.state.y_range
39 x_range.bounds = minrt, maxrt
40 y_range.bounds = minmz, maxmz
41
42
43points = hv.Points(
44 spectradf, kdims=["RT", "mz"], vdims=["inty"], label="MS1 survey scans"
45).opts(
46 fontsize={"title": 16, "labels": 14, "xticks": 6, "yticks": 12},
47 color=np.log(dim("int")),
48 colorbar=True,
49 cmap="Magma",
50 width=1000,
51 height=1000,
52 tools=["hover"],
53)
54
55raster = (
56 hd.rasterize(
57 points,
58 cmap=process_cmap("blues", provider="bokeh"),
59 aggregator=ds.sum("inty"),
60 cnorm="log",
61 alpha=10,
62 min_alpha=0,
63 )
64 .opts(active_tools=["box_zoom"], tools=["hover"], hooks=[new_bounds_hook])
65 .opts( # weird.. I have no idea why one has to do this. But with one opts you will get an error
66 plot=dict(
67 width=800,
68 height=800,
69 xlabel="Retention time (s)",
70 ylabel="mass/charge (Da)",
71 )
72 )
73)
74
75hd.dynspread(raster, threshold=0.7, how="add", shape="square")
Result:
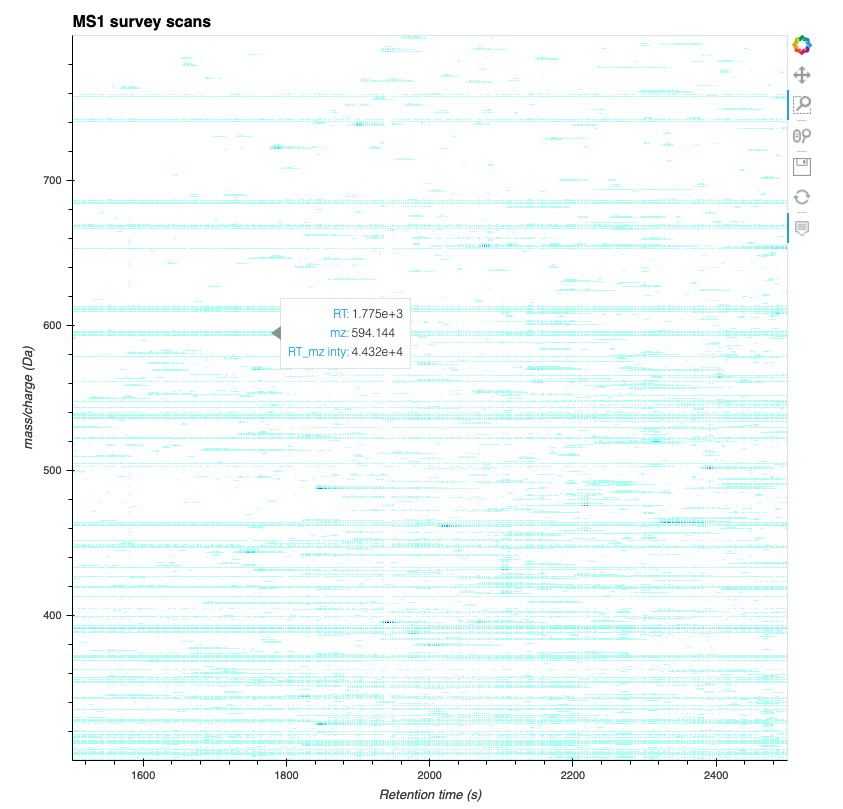
With this you can also easily create whole dashboards like the one hosted here on a Binder instance. If you are reading/executing this on Binder already, execute the next cell to get a link to your current instance.
1import os
2from IPython.display import Markdown as md
3
4md(
5 "When you are in binder already, you can quickly open the app [here]({}/msbokehapps).".format(
6 os.getenv("JUPYTERHUB_SERVICE_PREFIX")
7 )
8)