Feature Linking#
The pyOpenMS feature grouping algorithms group corresponding features (e.g., of same analyte) from multiple FeatureMap objects into a ConsensusMap.
Linking is primarily done based on spatial proximity (e.g., similar retention time and m/z).
It is, thus, advisable to perform a map alignment before features linking.
Optionally, identification data can be considered to prevent linking of features with different identifications.
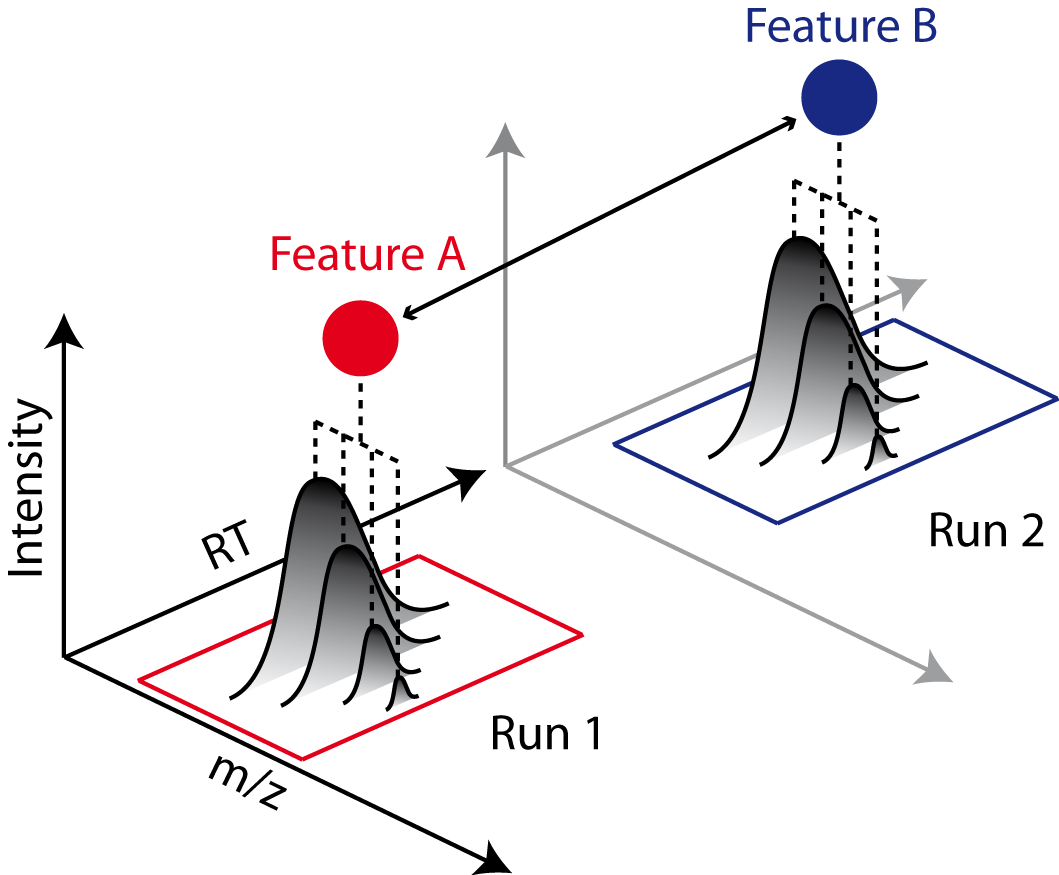
Different feature grouping algorithms with slightly different implementations are runtime characteristics are available in pyOpenMS:
We now perform a features linking using the FeatureGroupingAlgorithmQT algorithm.
Download Example Data#
import pyopenms as oms
from urllib.request import urlretrieve
base_url = (
"https://raw.githubusercontent.com/OpenMS/pyopenms-docs/master/src/data/"
)
feature_files = [
"BSA1_F1.featureXML",
"BSA2_F1.featureXML",
"BSA3_F1.featureXML",
]
feature_maps = []
# download the feature files and store feature maps in list (feature_maps)
for feature_file in feature_files:
urlretrieve(base_url + feature_file, feature_file)
feature_map = oms.FeatureMap()
oms.FeatureXMLFile().load(feature_file, feature_map)
feature_maps.append(feature_map)
features Linking Algorithm#
All FeatureMap objects will be combined in a ConsensusMap.
feature_grouper = oms.FeatureGroupingAlgorithmQT()
consensus_map = oms.ConsensusMap()
file_descriptions = consensus_map.getColumnHeaders()
# collect information about input maps
for i, feature_map in enumerate(feature_maps):
file_description = file_descriptions.get(i, oms.ColumnHeader())
file_description.filename = feature_map.getDataProcessing()[0].getMetaValue(
"parameter: in"
)[:-5]
file_description.size = feature_map.size()
file_description.unique_id = feature_map.getUniqueId()
file_descriptions[i] = file_description
consensus_map.setColumnHeaders(file_descriptions)
feature_grouper.group(feature_maps, consensus_map)