Spectrum Normalization#
Normalization by base peak intensity is a fundamental processing step in mass spectrometry. This method scales the peak intensities in a spectrum such that the highest peak reaches a maximum value, typically set to one. This approach facilitates the comparison of different spectra by standardizing the intensity scale.
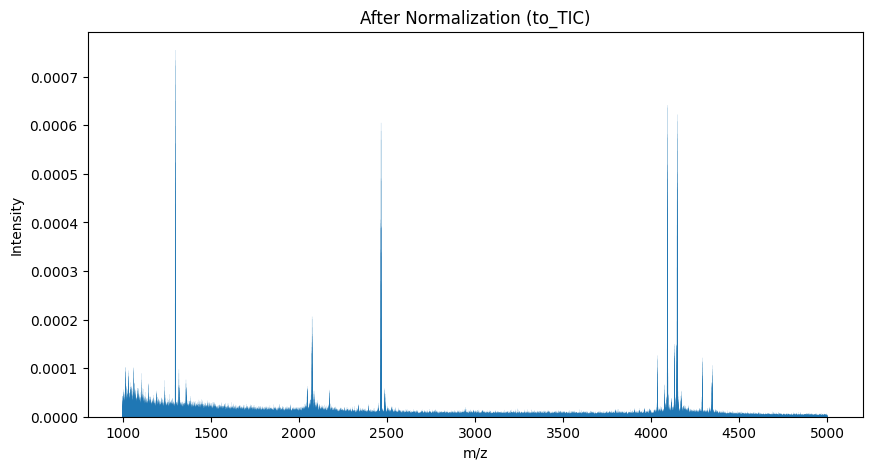
Loading the Raw Data#
To begin, we need to load the mass spectrometry data. The following Python code demonstrates how to load a spectrum from an mzML file using the pyOpenMS library.
1from urllib.request import urlretrieve
2import pyopenms as oms
3import matplotlib.pyplot as plt
4
5gh = "https://raw.githubusercontent.com/OpenMS/pyopenms-docs/master"
6urlretrieve(gh + "/src/data/peakpicker_tutorial_1_baseline_filtered.mzML", "tutorial.mzML")
7
8exp = oms.MSExperiment()
9oms.MzMLFile().load("tutorial.mzML", exp)
10
11plt.bar(exp.getSpectrum(0).get_peaks()[0], exp.getSpectrum(0).get_peaks()[1], snap=False)
12plt.show()
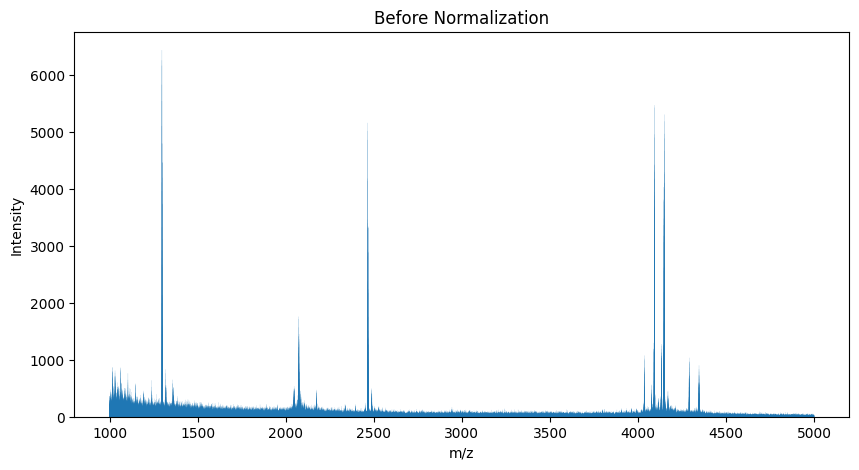
Normalization Procedure#
After loading the data, the next step is to apply normalization.
1normalizer = oms.Normalizer()
2param = normalizer.getParameters()
3param.setValue("method", "to_one")
4normalizer.setParameters(param)
5
6normalizer.filterPeakMap(exp)
7plt.bar(exp.getSpectrum(0).get_peaks()[0], exp.getSpectrum(0).get_peaks()[1], snap=False)
8plt.show()
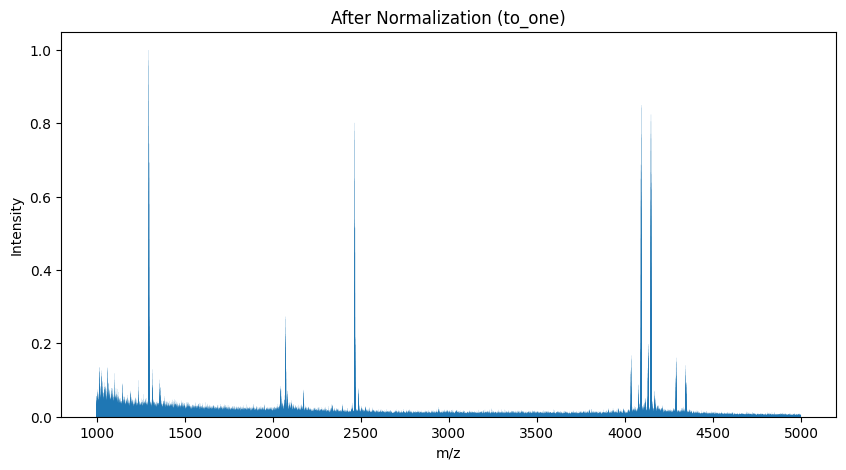
TIC Normalization#
Another approach to normalization is using the Total Ion Count (TIC). This method adjusts the intensities so that their total sum equals 1.0 in each mass spectrum.
1param.setValue("method", "to_TIC")
2normalizer.setParameters(param)
3normalizer.filterPeakMap(exp)
4plt.bar(exp.getSpectrum(0).get_peaks()[0], exp.getSpectrum(0).get_peaks()[1], snap=False)
5plt.show()