Peptide Search#
In MS-based proteomics, fragment ion mass spectra (MS2) are often interpreted by comparing them against a theoretical set of mass spectra generated from a FASTA database. OpenMS contains a (simple) implementation of such a “search engine” that compares experimental against theoretical mass spectra generated from an enzymatic or chemical digest of a proteome (e.g. tryptic digest).
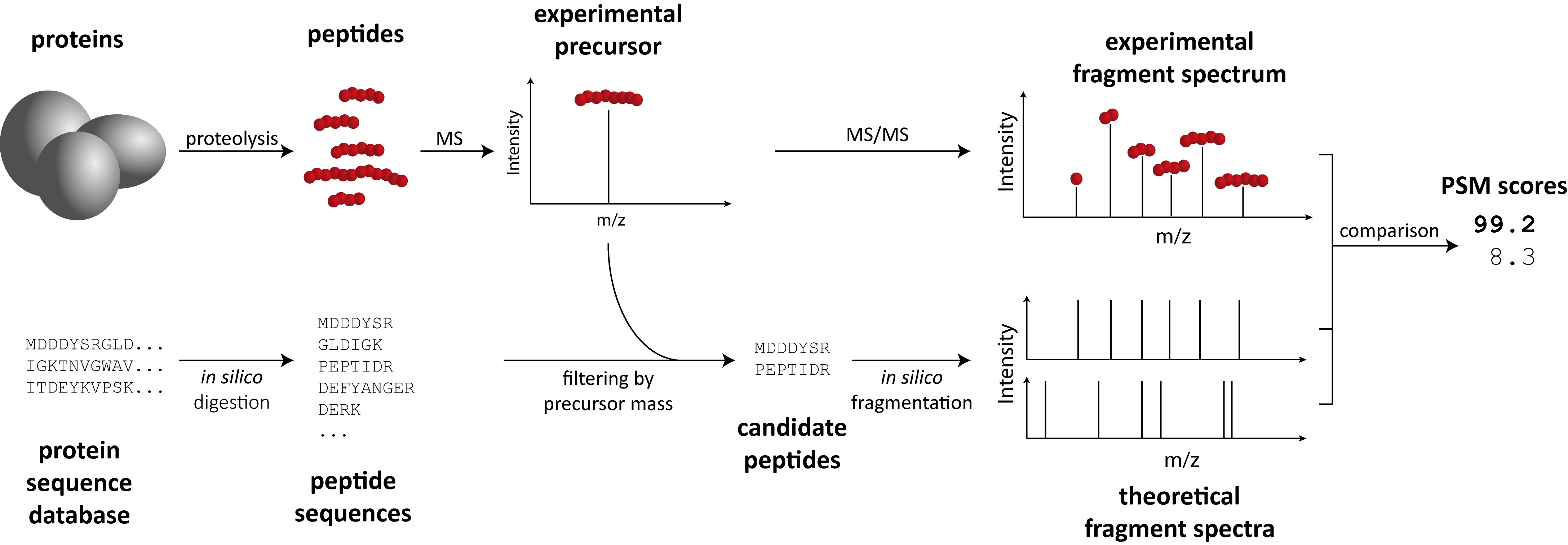
SimpleSearch#
In most proteomics applications, a dedicated search engine (such as Comet,
Crux, Mascot, MSGFPlus, MSFragger, Myrimatch, OMSSA, SpectraST or XTandem;
all of which are supported by pyOpenMS) will be used to search data. Here, we will
use the internal SimpleSearchEngineAlgorithm from OpenMS used for teaching
purposes. This makes it very easy to search an (experimental) mzML file against
a fasta database of protein sequences:
1from urllib.request import urlretrieve
2import pyopenms as oms
3
4gh = "https://raw.githubusercontent.com/OpenMS/pyopenms-docs/master"
5urlretrieve(gh + "/src/data/SimpleSearchEngine_1.mzML", "searchfile.mzML")
6urlretrieve(gh + "/src/data/SimpleSearchEngine_1.fasta", "search.fasta")
7protein_ids = []
8peptide_ids = []
9oms.SimpleSearchEngineAlgorithm().search(
10 "searchfile.mzML", "search.fasta", protein_ids, peptide_ids
11)
This will print search engine output including the number of peptides and proteins in the database and how many mass spectra were matched to peptides and proteins:
Peptide statistics
unmatched : 0 (0 %)
target/decoy:
match to target DB only: 3 (100 %)
match to decoy DB only : 0 (0 %)
match to both : 0 (0 %)
Peptide-Spectrum Match (PSM) Inspection#
We can now investigate the individual hits as we have done before in the identification tutorial.
1for peptide_id in peptide_ids:
2 # Peptide identification values
3 print(35 * "=")
4 print("Peptide ID m/z:", peptide_id.getMZ())
5 print("Peptide ID rt:", peptide_id.getRT())
6 print("Peptide scan index:", peptide_id.getMetaValue("scan_index"))
7 print("Peptide scan name:", peptide_id.getMetaValue("scan_index"))
8 print("Peptide ID score type:", peptide_id.getScoreType())
9 # PeptideHits
10 for hit in peptide_id.getHits():
11 print(" - Peptide hit rank:", hit.getRank())
12 print(" - Peptide hit charge:", hit.getCharge())
13 print(" - Peptide hit sequence:", hit.getSequence())
14 mz = (
15 hit.getSequence().getMonoWeight(
16 oms.Residue.ResidueType.Full, hit.getCharge()
17 )
18 / hit.getCharge()
19 )
20 print(" - Peptide hit monoisotopic m/z:", mz)
21 print(
22 " - Peptide ppm error:", abs(mz - peptide_id.getMZ()) / mz * 10**6
23 )
24 print(" - Peptide hit score:", hit.getScore())
We notice that the second PSM was found for the third
term:mass spectrum in the file for a precursor at \(775.38\) m/z for the sequence
RPGADSDIGGFGGLFDLAQAGFR.
1tsg = oms.TheoreticalSpectrumGenerator()
2thspec = oms.MSSpectrum()
3p = oms.Param()
4p.setValue("add_metainfo", "true")
5tsg.setParameters(p)
6peptide = oms.AASequence.fromString("RPGADSDIGGFGGLFDLAQAGFR")
7tsg.getSpectrum(thspec, peptide, 1, 1)
8# Iterate over annotated ions and their masses
9for ion, peak in zip(thspec.getStringDataArrays()[0], thspec):
10 print(ion, peak.getMZ())
11
12e = oms.MSExperiment()
13oms.MzMLFile().load("searchfile.mzML", e)
14print("Spectrum native id", e[2].getNativeID())
15mz, i = e[2].get_peaks()
16peaks = [(mz, i) for mz, i in zip(mz, i) if i > 1500 and mz > 300]
17for peak in peaks:
18 print(peak[0], "mz", peak[1], "int")
Comparing the theoretical and the experimental mass spectrum for
RPGADSDIGGFGGLFDLAQAGFR we can easily see that the most abundant ions in are
\(\ce{y8}\) (\(\ce{877.452}\) m/z), \(\ce{b10}\) (\(926.432\)), \(\ce{y9}\)
(\(1024.522\) m/z) and \(\ce{b13}\) (\(1187.544\) m/z).
Visualization#
When loading the searchfile.mzML into the OpenMS
visualization software TOPPView, we can convince ourselves that the observed
mass spectrum indeed was generated by the peptide RPGADSDIGGFGGLFDLAQAGFR by loading
the corresponding theoretical mass spectrum into the viewer using “Tools”->”Generate
theoretical spectrum”:
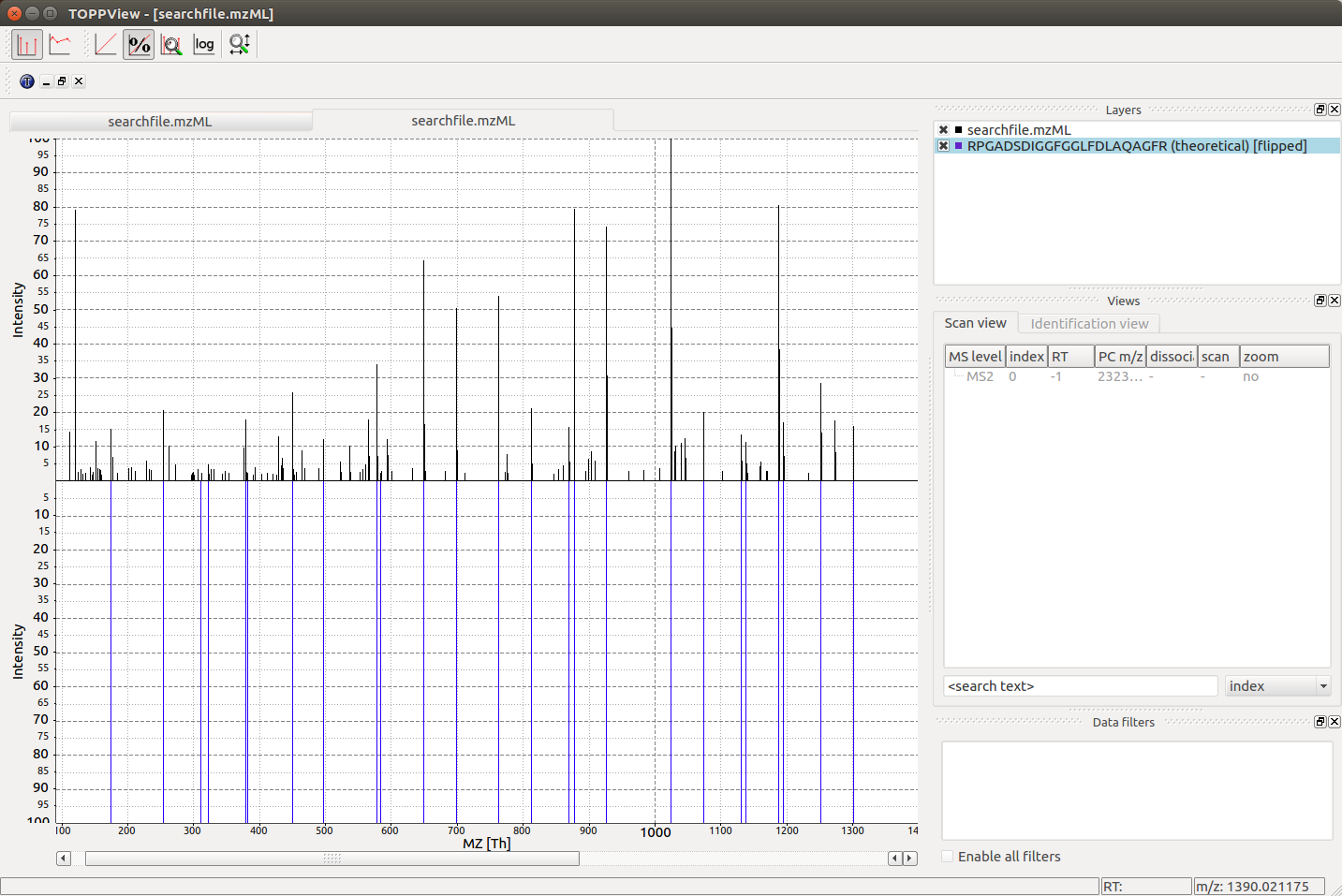
From our output above, we notice that the second PSM
at \(775.38\) m/z for sequence RPGADSDIGGFGGLFDLAQAGFR was found with an error
tolerance of \(2.25\ ppm\), therefore if we set the precursor mass tolerance to \(4\
ppm\ (\pm 2\ ppm)\), we expect that we will not find the hit at \(775.38\) m/z any more:
1salgo = oms.SimpleSearchEngineAlgorithm()
2p = salgo.getDefaults()
3print(p.items())
4p[b"precursor:mass_tolerance"] = 4.0
5salgo.setParameters(p)
6
7protein_ids = []
8peptide_ids = []
9salgo.search("searchfile.mzML", "search.fasta", protein_ids, peptide_ids)
10print("Found", len(peptide_ids), "peptides")
As we can see, using a smaller precursor mass tolerance leads the algorithm to
find only one hit instead of two. Similarly, if we use the wrong enzyme for
the digestion (e.g. p[b'enzyme'] = "Formic_acid"), we find no results.
More detailed example#
Now include some additional decoy database generation step as well as subsequent FDR filtering.
1from urllib.request import urlretrieve
2
3searchfile = "../../../src/data/BSA1.mzML"
4searchdb = "../../../src/data/18Protein_SoCe_Tr_detergents_trace.fasta"
5
6# generate a protein database with additional decoy sequenes
7targets = list()
8decoys = list()
9oms.FASTAFile().load(
10 searchdb, targets
11) # read FASTA file into a list of FASTAEntrys
12decoy_generator = oms.DecoyGenerator()
13for entry in targets:
14 rev_entry = oms.FASTAEntry(entry) # copy entry
15 rev_entry.identifier = "DECOY_" + rev_entry.identifier # mark as decoy
16 aas = oms.AASequence().fromString(
17 rev_entry.sequence
18 ) # convert string into amino acid sequence
19 rev_entry.sequence = decoy_generator.reverseProtein(
20 aas
21 ).toString() # reverse
22 decoys.append(rev_entry)
23
24target_decoy_database = "search_td.fasta"
25oms.FASTAFile().store(
26 target_decoy_database, targets + decoys
27) # store the database with appended decoy sequences
28
29# Run SimpleSearchAlgorithm, store protein and peptide ids
30protein_ids = []
31peptide_ids = []
32
33# set some custom search parameters
34simplesearch = oms.SimpleSearchEngineAlgorithm()
35params = simplesearch.getDefaults()
36score_annot = [b"fragment_mz_error_median_ppm", b"precursor_mz_error_ppm"]
37params.setValue(b"annotate:PSM", score_annot)
38params.setValue(b"peptide:max_size", 30)
39simplesearch.setParameters(params)
40
41simplesearch.search(searchfile, target_decoy_database, protein_ids, peptide_ids)
42
43# Annotate q-value
44oms.FalseDiscoveryRate().apply(peptide_ids)
45
46# Filter by 1% PSM FDR (q-value < 0.01)
47idfilter = oms.IDFilter()
48idfilter.filterHitsByScore(peptide_ids, 0.01)
49idfilter.removeDecoyHits(peptide_ids)
50
51# store PSM-FDR filtered
52oms.IdXMLFile().store(
53 "searchfile_results_1perc_FDR.idXML", protein_ids, peptide_ids
54)
However, usually researchers are interested in the most confidently identified proteins. This so called protein inference problem is a difficult problem because of often occurring shared/ambiguous peptides. To be able to calculate a target/decoy-based FDR on the protein level, we need to assign scores to proteins first (e.g. based on their observed peptides). This is done by applying one of the available protein inference algorithms on the peptide and protein IDs.
1protein_ids = []
2peptide_ids = []
3
4# Re-run search since we need to keep decoy hits for inference
5simplesearch.search(searchfile, target_decoy_database, protein_ids, peptide_ids)
6
7# Run inference
8bpia = oms.BasicProteinInferenceAlgorithm()
9params = bpia.getDefaults()
10# FDR with groups currently not supported in pyopenms
11params.setValue("annotate_indistinguishable_groups", "false")
12bpia.setParameters(params)
13bpia.run(peptide_ids, protein_ids)
14
15
16# Annotate q-value on protein level
17# Removes decoys in default settings
18oms.FalseDiscoveryRate().apply(protein_ids)
19
20# Filter targets by 1% protein FDR (q-value < 0.01)
21idfilter = oms.IDFilter()
22idfilter.filterHitsByScore(protein_ids, 0.01)
23
24# Restore valid references into the proteins
25remove_peptides_without_reference = True
26idfilter.updateProteinReferences(
27 peptide_ids, protein_ids, remove_peptides_without_reference
28)
29
30# store protein-FDR filtered
31oms.IdXMLFile().store(
32 "searchfile_results_1perc_protFDR.idXML", protein_ids, peptide_ids
33)